Circulating Tumor DNA: Overview and Application
Circulating tumor DNA has emerged as one of the most powerful biomarkers in modern oncology, enabling non-invasive, real-time assessment of tumor biology.

Background
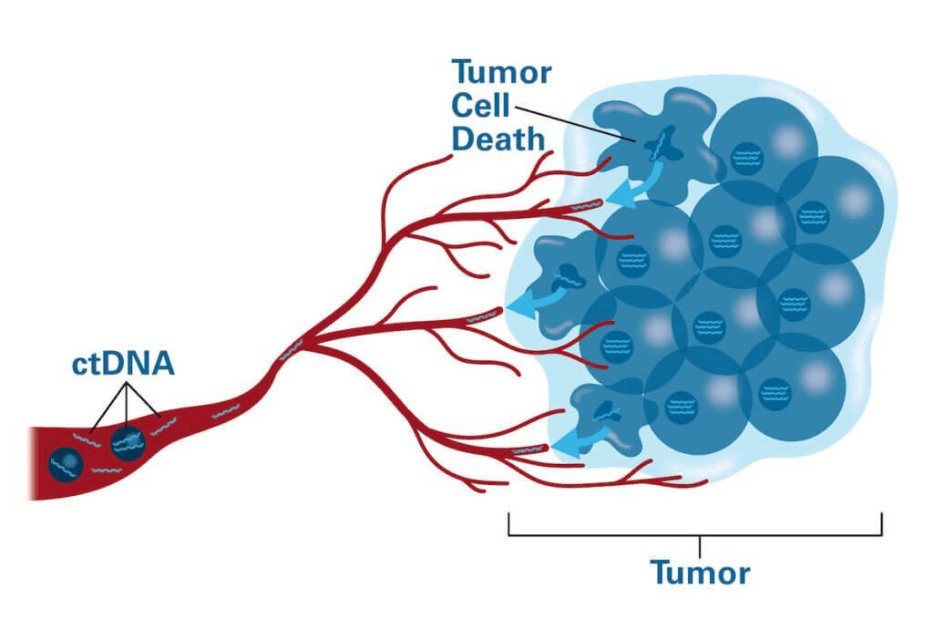
Circulating tumor DNA (ctDNA) represents short DNA fragments released into the peripheral blood by malignant cells. ctDNA is a component of cell-free DNA (cfDNA), which also includes DNA shed from healthy tissues. ctDNA gained clinical relevance with advances in next-generation sequencing (NGS) and digital PCR, enabling detection of tumor-specific genetic alterations at very low variant allele frequencies (VAFs).
ctDNA analysis, often termed liquid biopsy, offers a non-invasive alternative to traditional tissue biopsy, allowing dynamic, real-time molecular profiling of cancer. ctDNA has potential applications across the entire cancer continuum, including early detection, molecular diagnostics, treatment selection, minimal residual disease (MRD) assessment, and relapse prediction.
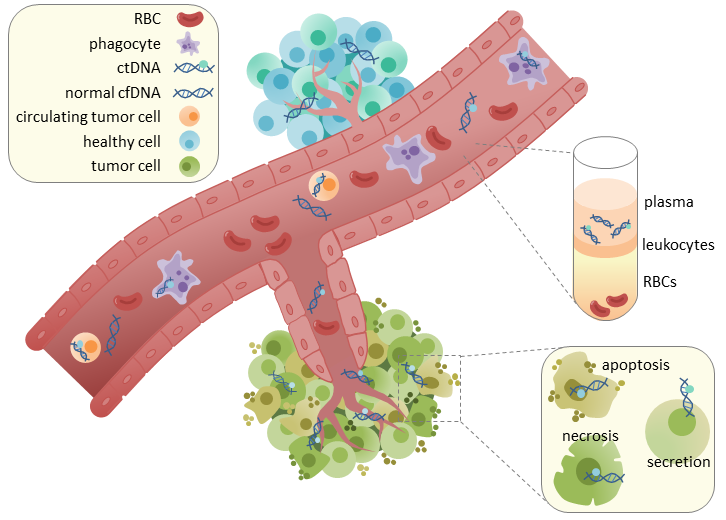
Biological Basis of ctDNA
Source and Release Mechanisms
ctDNA enters the bloodstream through:
- Apoptosis: orderly DNA fragmentation during programmed cell death
- Necrosis: uncontrolled release of DNA from dying cells in poorly vascularized tumor areas
- Active secretion: DNA-bound extracellular vesicles, exosomes, and microvesicles
- Circulating Tumor Cells (CTCs): CTC lysis contributes small amounts of DNA
Tumor Shedding Variability
ctDNA abundance differs by cancer type:
- High-shedding tumors: lung, colorectal, breast, high-grade serous ovarian cancer
- Low-shedding tumors: gliomas (due to blood-brain barrier), indolent hematologic malignancies, early-stage disease
Analytical Platforms
- Digital PCR (dPCR/ddPCR)
- Extremely high sensitivity for known hotspot mutations
- Ideal for targeted assays (e.g., EGFR T790M)
- Limited throughput and breadth
- Next-Generation Sequencing (NGS)
- Includes targeted panels, hybrid-capture sequencing, and whole-genome/exome sequencing
- Broad mutation detection (SNVs, indels, CNAs, fusions)
- Can incorporate error-corrected molecular barcodes to enhance sensitivity (down to 0.01% VAF)
- Tumor-Informed Assays
- Examples: Signatera, Safe-Seq
- Based on sequencing the patient’s tumor to identify unique mutations
- Ultra-sensitive MRD detection
- High specificity with low false-positive rates
- Tumor-Agnostic Assays
- Examples: Guardant Reveal, GRAIL
- Detect common cancer mutations and methylation patterns without a patient-specific tumor sample
- Broader applicability but often slightly less sensitive for MRD
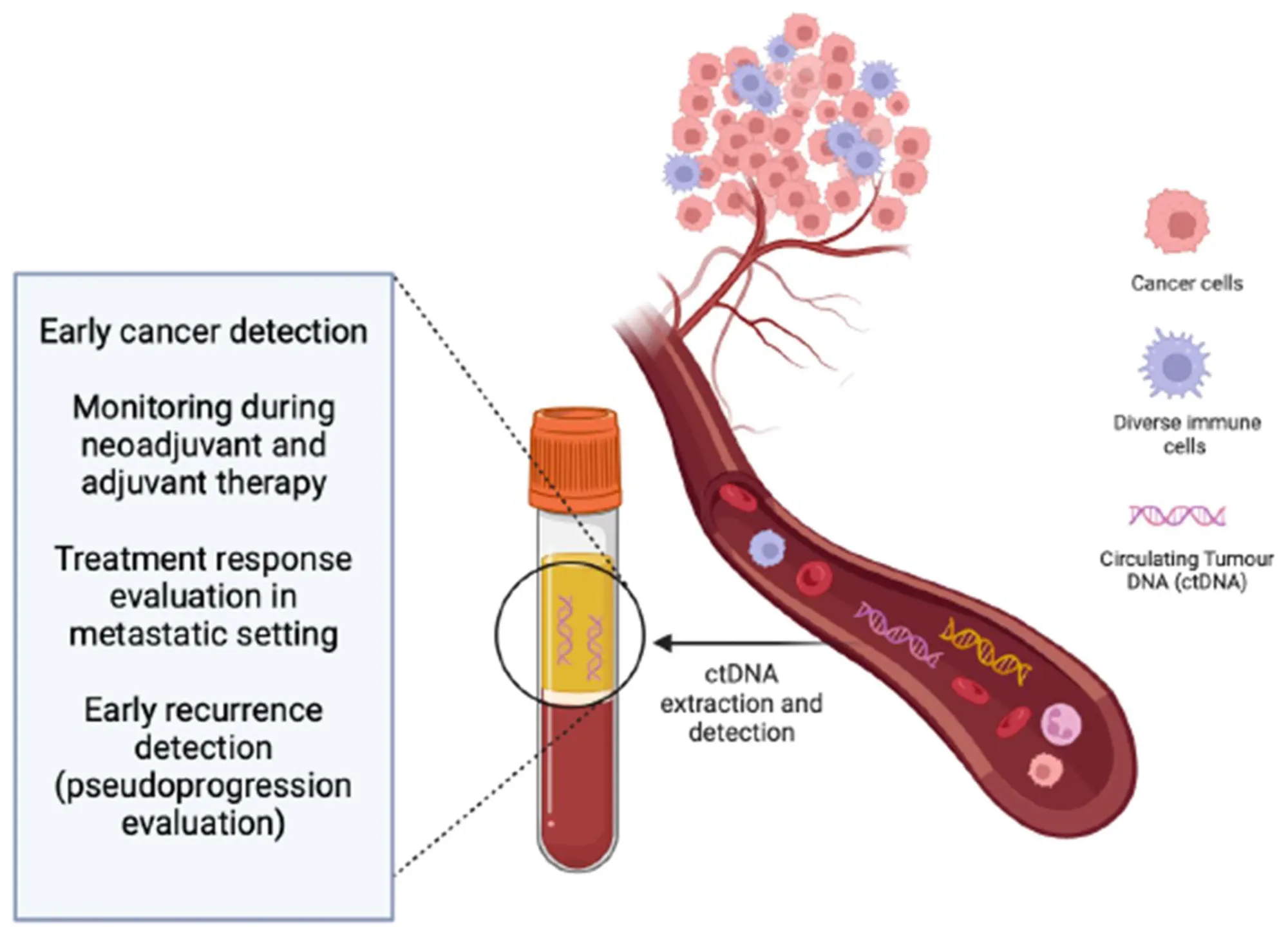
Clinical Applications of ctDNA
- Molecular Profiling for Treatment Selection
- ctDNA enables rapid molecular characterization of tumors when tissue is insufficient, inaccessible, or outdated
- Examples Across Tumor Types:
- NSCLC: EGFR (L858R, exon 19 del), ALK/ROS1 fusions, MET exon 14 skipping, KRAS, resistance mutations
- Colorectal cancer (CRC): KRAS & BRAF mutations
- Breast cancer: ESR1 mutations conferring endocrine therapy resistance
- Prostate cancer: BRCA1/2, ATM, HRD genes guiding PARP inhibitor use
- Liquid biopsy is endorsed by major guidelines (NCCN, ASCO, ESMO) for genomic testing across many solid tumors
- Monitoring Treatment Response
- Because ctDNA has a short half-life, changes in ctDNA levels often precede radiologic responses
- Early clearance can correlate with stronger progression-free and overall survival
- Persistent ctDNA during treatment may signal inadequate response or resistance
- Immunotherapy: ctDNA decline may associate with benefit and long-term survival
- This dynamic monitoring is increasingly used in clinical trials
- Minimal Residual Disease (MRD) Detection
- Rationale: After curative-intent treatment (surgery or chemoradiation), small numbers of residual cancer cells may persist undetected by imaging
- ctDNA can detect molecular relapse months before clinical recurrence
- Clinical Impact:
- Escalation: treat MRD-positive patients earlier
- De-escalation: potentially avoid chemotherapy in MRD-negative patients
- Surveillance: molecular monitoring offers earlier warning of relapse
- Early Cancer Detection and Screening
- Multi-cancer early detection (MCED) tests use ctDNA mutation signatures, fragmentomics, and methylation patterns
- Studies show >90% specificity, but variable sensitivity, especially for stage I disease in specific cancer types
- The main challenges are low ctDNA shedding in early disease and interference from clonal hematopoiesis
Emerging Research and Future Directions
- Multi-omic Liquid Biopsy
- Combining:
- ctDNA mutations
- Methylation profiling
- Fragmentomics
- Cell-free RNA
- Proteomics
- This approach aims to improve early detection and biological resolution
- Combining:
- ctDNA as a Surrogate Endpoint
- Regulatory agencies and trial sponsors are exploring ctDNA as:
- An early marker of therapeutic efficacy
- A surrogate for relapse-free or overall survival
- A tool for rapid go/no-go decisions in clinical development
- Regulatory agencies and trial sponsors are exploring ctDNA as:
- AI-Enhanced Analysis
- Machine learning applied to methylation and fragmentation patterns dramatically improves detection accuracy
- Integration Into Standard Care
- Future oncology pathways will likely use ctDNA for:
- Initial genomic profiling
- MRD-guided treatment
- Real-time monitoring
- Identifying escape pathways and resistance
- Molecular confirmation of recurrence
- Future oncology pathways will likely use ctDNA for:

Conclusion
Circulating tumor DNA has emerged as one of the most powerful biomarkers in modern oncology, enabling non-invasive, real-time assessment of tumor biology. Its applications span molecular diagnosis, therapy selection, treatment response monitoring, MRD detection, and early relapse prediction. Although challenges remain, including assay variability, limited sensitivity in early-stage disease, and high costs, the evidence supporting ctDNA-guided clinical decision-making continues to grow rapidly.
As multi-omic technologies mature and prospective clinical trials validate MRD-guided therapeutic strategies, ctDNA is poised to transform oncology practice by personalizing treatment intensity, improving early detection, and enabling dynamic disease monitoring. Ultimately, ctDNA holds the promise of more precise, proactive, and biologically driven cancer care.

References
- Wan JCM et al. Nature Reviews Cancer. 2017.
- Cohen JD et al. Science. 2018.
- Abbosh C et al. TRACERx study. New England Journal of Medicine. 2017.
- Reinert T et al. GALAXY/CIRCULATE-Japan. Nature Medicine. 2022.
- Maron SB et al. JCO Precision Oncology. 2021.
- Tie J et al. DYNAMIC trial. New England Journal of Medicine. 2022.
- McDonald BR et al. Cancer Discovery. 2019.
- Parikh AR et al. JAMA Oncology. 2021.
- Newman AM et al. Nature Biotechnology. 2016.
- Merker JD et al. ASCO/AMP guidelines. Journal of Clinical Oncology. 2018.
*Information presented on RxTeach does not represent the opinion of any specific company, organization, or team other than the authors themselves. No patient-provider relationship is created.

