Bugs, Drugs, and Gram-Negative Resistance
The second most common reason for emergency department (ED) visits is acute upper respiratory infections with approximately 5.8 million patients being treated and discharged from the ED.

The background section of this article will provide a very general overview of antibiotics, how they are utilized, and why appropriate use is vital. If you are well familiar with antibiotics and their respective bugs, it might be worth skipping to the clinical pearls section below which focuses on beta-lactamase resistance mechanisms in Gram-negative bacteria. A free case study is attached!
Background:
The second most common reason for emergency department (ED) visits is acute upper respiratory infections with approximately 5.8 million patients being treated and discharged from the ED. This data is from a 2018 survey completed by the Agency for Healthcare Research and Quality. Other common reasons for ED visits include urinary tract and skin and soft tissue infections.
Most often, these types of acute infections are caused by bacteria, and antibiotics are used to treat them. "Anti", meaning kill, and “biotics”, meaning relating to or resulting from living things. Antibiotics work through a number of mechanisms, but generally they either inhibit the growth of or destroy bacteria. Antibiotics are great for eradicating pathogenic bacteria; however, antibiotics are not selective and can also kill synergistic bacteria in the human body.
There are trillions of bacteria living on and inside your body right now. They live in symbiosis with you to aid in digestion, disease prevention, nutrient absorption, and other normal bodily functions. When antibiotics are prescribed, they can disrupt our normal flora by killing off “good” bacteria. Eliminating too many “good” bacteria can reduce competition for other (possibly pathogenic) bacteria to proliferate and cause new infections. To maximize the effect of antibiotics and minimize the disruption of normal flora, the most narrow antibiotics are prescribed for the shortest effective duration. If antibiotics are prescribed for an extended duration, bacteria can develop resistance.
The first antibiotic was discovered in 1928 by the great Alexander Flemming. He was studying staphylococcal bacteria when he noticed an uncovered Petri dish contaminated with mold spores. He then observed the bacteria nearest the mold colonies beginning to die, thus leading to the accidental discovery of penicillin.
Penicillin is still the drug-of-choice for a select number of infections today (e.g., syphilis and sensitive streptococci infections). The widespread use of penicillin began around 1942. Only five years later occurred the first documented case of bacterial resistance to penicillin. Antibiotic-resistant organisms have become a major concern over the last few decades. More than 2.8 million antibiotic-resistant infections occur in the United States each year. As a result, more than 35,000 people die. When Clostridioides difficile (a bacterium that is not typically resistant, but can cause deadly diarrhea and is associated with antibiotic use) is added, the US exceeds 3 million infections and 48,000 deaths.
To combat the growing resistance, the Centers for Disease Control (CDC) has started an Antibiotic Resistance Solution Initiative. This is one of many groups formed to help fight bacterial resistance. Not only does microbial resistance incur a morbidity and mortality burden, but it also raises the matter of untreatable super bugs.
A “super bug” is a strain of bacteria that has become resistant to most, if not all, available antibiotics. If a patient is infected with a bacterium that cannot be treated with available antibiotics, the infection likely cannot be cured, and the patient is most likely going to die. Without antimicrobial stewardship (AMS) and frugal use of antibiotics, the world could fall victim to another pandemic similar to the Black Death (caused by the bacterium Yersinia pestis). The focus of AMS is to avoid unnecessary coverage, minimize excessive durations, and de-escalate therapy as soon as possible.
When a patient is admitted with an unknown infection, empiric antibiotic therapy is started. Empiric therapy typically consists of broad antibiotic coverage that will ensure appropriate therapy until the pathogen is identified. Once cultures are sent, the team waits for a Gram stain and culture to result. From this data, definitive therapy can be started. Definitive therapy refers to treatment with a narrow-spectrum antibiotic once the causative bacterium is known. Narrow refers to an antibiotic that covers a minimal number of pathogens while broad coverage would be an antibiotic that covers a wide range of organisms.
In addition to de-escalating from empiric to definitive therapy, the source of infection is also pertinent to AMS. The source will guide practitioners on how long to prescribe the antibiotic to minimize the chance of resistance. Many healthcare systems have an AMS committee which includes physicians, pharmacists, nurses, and other team members. An AMS committee can help monitor the use of antibiotics in the hospital by assessing appropriateness, duration, route, and cost. When a physician suspects a bacterial infection, one of the first steps is to culture the infection site. This is typically done prior to the initiation of antibiotics. When a culture is collected, the microbiology lab will perform a Gram stain. A Gram stain is a laboratory technique used to identify if bacteria are present at the infection site and can help guide physicians in prescribing treatment.
If present, the bacteria will stain either purple/blue or pink/red. This is dependent on the presence of a peptidoglycan layer in the bacteria. Gram-positive (GP) bacteria will stain purple/blue as the bacteria contain a thick cell wall of peptidoglycan. Gram-negative (GN) bacteria will stain pink/red as the bacteria only contain a cell membrane.
GP and GN bacteria differ in the mechanisms of resistance they use against antibiotics. The main mechanisms of resistance for GP bacteria include the production of beta-lactamases or altering the structure of the target site, penicillin-binding protein (PBP). Beta-lactamases will cause enzymatic degradation of the antibiotics, rendering the medication inactive against the bacteria. Altering the PBP target site will decrease the affinity and binding of the antibiotic to the bacterial component. If the antibiotic cannot bind to the bacteria, it will not kill the bacteria.
To determine resistance, there are rapid microbial identification (ID) and antibiotic susceptibility testing (AST) instruments that can report specific genes, phenotypes, and breakpoints that the bacteria express. These instruments can provide results much faster than a culture and sensitivity (C&S) report. They are a great resource for AMS teams to de-escalate to definitive antibiotic therapy in clinical practice. One of the genes detected for GP organisms is the vanA gene. This can be expressed by the Staphylococcus and Enterococcus species. The presence of the vanA gene will result in a decreased binding affinity of vancomycin to the target site on the bacterial cell wall, therefore creating resistance.
The mechanisms of GN resistance are more complicated. There are three main categories: intrinsic, acquired, and adaptive. Intrinsic resistance is when the bacteria contain genes that encode antimicrobial properties. For example, Enterococcus spp. has intrinsic resistance against gentamicin, an aminoglycoside that interferes with bacterial protein synthesis by binding to a target inside the cell. Enterococcus spp. has a thick cell wall layer that gentamicin cannot penetrate. This is why aminoglycosides are also used in combination with other antibiotics that are active against GP organisms, such as vancomycin, for GP infections.
Acquired resistance occurs when the bacteria are subjected to selective pressure. Selective pressure will kill susceptible bacteria and allow antibiotic-resistant organisms to survive and multiply. Acquired resistance mechanisms include beta-lactamase production, diminished cell membrane permeability, changes in PBPs, loss of porins, presence of antibiotic-modifying enzymes, target site mutations, and increased efflux pump expression. Adaptive resistance is recognized as inducible and occurs under the presence of specific antibiotics and environmental stressors.
Clinical Pearls:
The focus of the clinical pearls for this article will be on the beta-lactamase resistance mechanisms in GN bacteria. Please refer to the attached PDF for a visual representation of the following information.
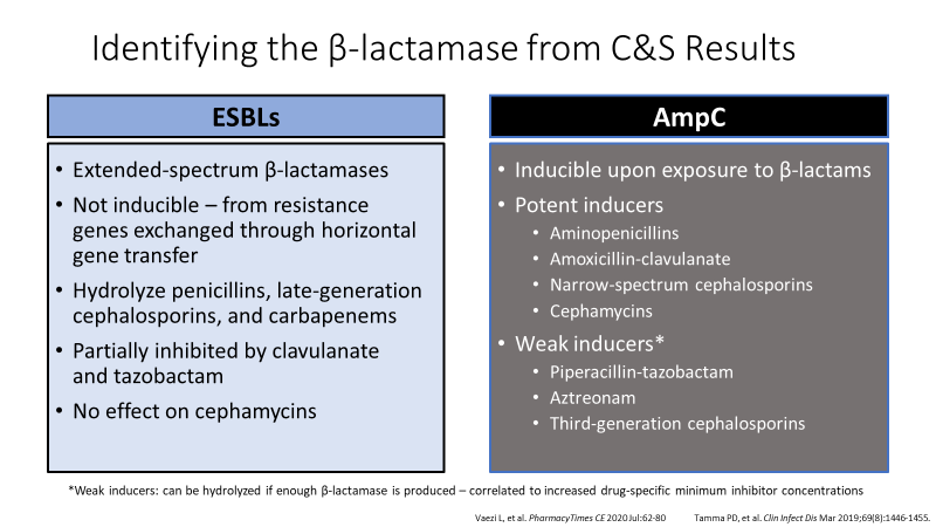
Many times in practice, a C&S results with a GN organism that is known to be a beta-lactamase producer. There are several Ambler classes to better categorize the different types of beta-lactamase producing organisms. For the purpose of this clinical pearl, we will focus on the Extended Spectrum Beta-Lactamase (ESBL) producers in Ambler Class A and the AmpC producers in Ambler Class C.
Common ESBL organisms: Klebsiella pneumonia, Klebsiella oxytoca, Escherichia coli, Proteus mirabilis
Common AmpC organisms: Enterobacter spp., Citrobacter freundii, Klebsiella aerogenes, Serratia marcescens
If an organism produces a beta-lactamase, the sensitivities will usually show a variety of “resistant” results to the antibiotics tested in the lab. Some providers will immediately jump to meropenem after seeing the results of a C&S with multiple resistant readings. However, the patterns of the resistant results on the C&S can help guide de-escalation to a narrower and more appropriate antibiotic. There are three steps to follow when determining what beta-lactamase producers the GN may be.
First, we must identify the differences between an ESBL and an AmpC organism. AmpC producers are known for their inducible resistance when exposed to specific antibiotics (see picture below).
Next, we need to analyze the C&S results (see attached PDF above):
- What organism is growing? Enterobacter cloacae. This organism is a known AmpC producer.
- Is the organism “resistant” to amoxicillin/clavulanic acid and cefoxitin? This is indicative of an AmpC producer.
- Is the organism “sensitive” or “intermediate” to amoxicillin/clavulanic acid and cefoxitin? This is indicative of an ESBL producer.
Finally, after analyzing the C&S and identifying if the pathogen is most likely an AmpC, we can take the following route of therapy de-escalation (see attached PDF above):
- Ceftriaxone: If too much beta-lactamase is produced, Enterobacter spp. can hydrolyze ceftriaxone and become resistant leaving the patient inadequately treated. Not a great option for AmpC producers.
- Meropenem: Stable against AmpC producers, but provides broad, unnecessary coverage. This can create selective pressures and future ESBL organisms. Not the most appropriate option.
- Cefepime: Weak inducer of the AmpC gene and contains a stable acyl-enzyme complex to withstand hydrolysis by AmpC beta-lactamases. This is the most narrow, appropriate option!
After a little critical thinking and analysis of C&S results, you can practice AMS principles in your clinical setting. Reserving carbapenems for true ESBLs and multi-drug resistant organisms (MDROs) is important to avoid increasing resistance rates and ensure we have effective antibiotics.
Overall, antimicrobial resistance is a complicated subject. There are many nuances and rabbit holes to go down in the literature. To guide practitioners in AMS practices, the Infectious Disease Society of America has recently released two guidance documents on the treatment of antimicrobial resistance in GN infections (Version 1.0) (Version 2.0). Save these documents and help practice AMS at your practice site!

